Sanjay Tyagi, Ph.D.
Professor of Medicine
sanjay.tyagi@rutgers.edu | Laboratory Website
+1-973-854-3372
room W310-E
NEWS
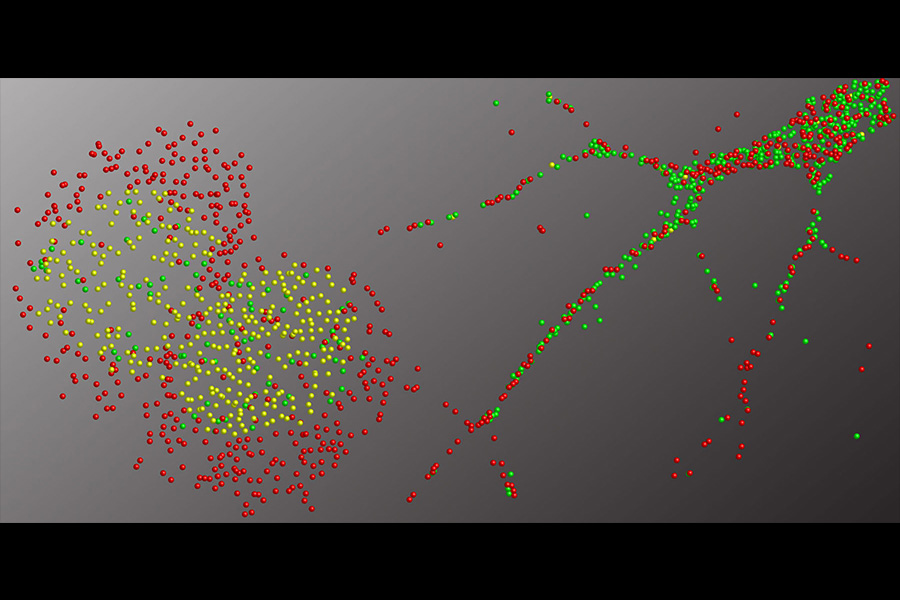
 The Journal of Immunology selected our paper for their cover. Colors of the rendered cells display density of different immune cell types in a tuberculosis granuloma based on their cytokine mRNA expression. Granulomas form as infected cells secrete cytokines to attract more immune cells to surround and isolate the site of infection.
The Journal of Immunology selected our paper for their cover. Colors of the rendered cells display density of different immune cell types in a tuberculosis granuloma based on their cytokine mRNA expression. Granulomas form as infected cells secrete cytokines to attract more immune cells to surround and isolate the site of infection.
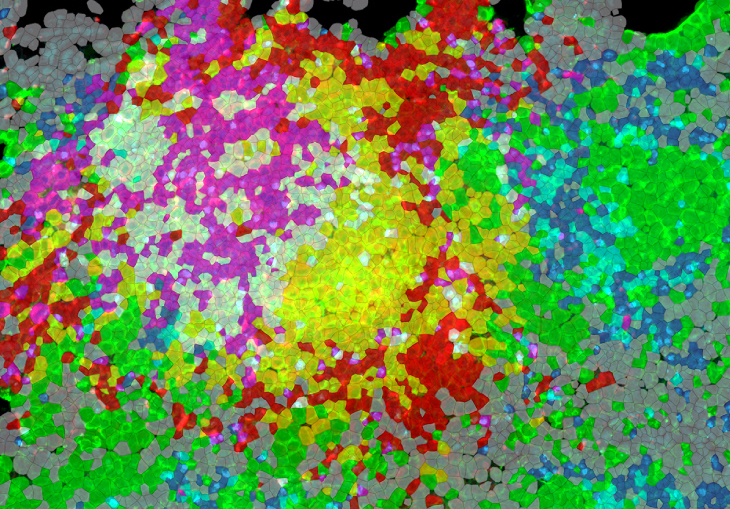
Ranjeet Kumar, Afsal Kolloli and colleagues describe their single-molecule FISH approach for imaging the architecture of tuberculosis granulomas in rabbits and macaques (www.aai.org). Granulomas are created by immune cells around mycobacteria to limit both bacterial dissemination and inflammation in the lung. The scientists classified immune cells based on mRNA expressions and mapped their distributions. This approach paves the way for more comprehensive spatial transcriptomic studies that will produce better understanding of the Tuberculosis disease.
Probes for RNA Imaging. We developed two popular probe chemistries for visualization of RNAs in situ: molecular beacon probes, that become fluorescent upon binding to target RNAs and thereby allow visualization of RNA dynamics and trafficking in live cells and single molecule FISH (Stellaris) probes, that allow visualization of single RNA molecules in cells after fixation.
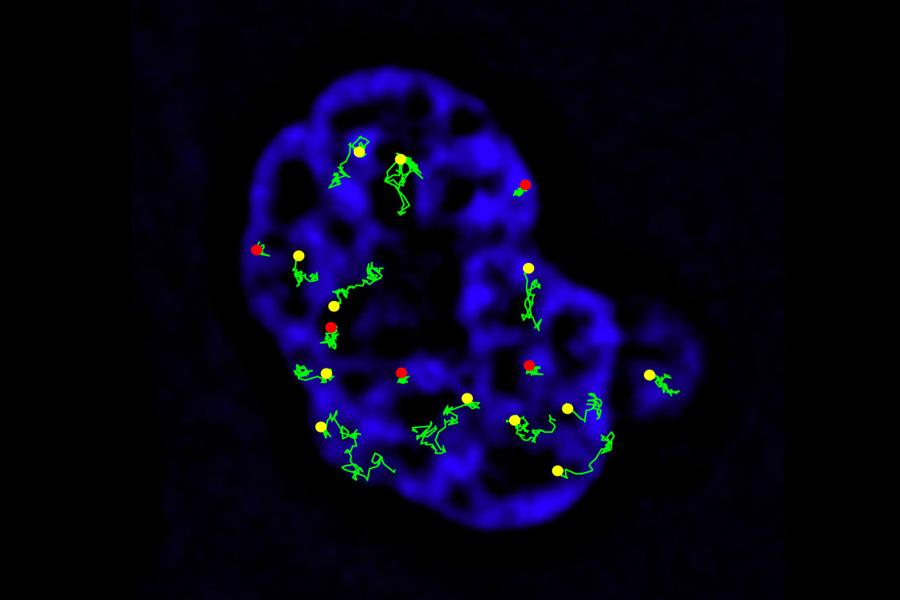
mRNA Traffic in Nucleus. How the large complex formed by mRNA and the proteins that bind to it during its synthesis, is able to travel from the site of transcription to the nuclear pores through the extremely dense chromatin has been an enigma. We visualized the transport of individual mRNA molecules soon after their synthesis at the gene locus. The analysis of the molecular tracks revealed that mRNP complexes explore the volume of the nucleus by simple Brownian diffusion. The motion of mRNP complexes is restricted to the interchromatin spaces. When the complexes drift into the dense chromatin, they tend to become immobilized. This glimpse into the dynamic architecture of the nucleus reveals a key step in the expression of genes.
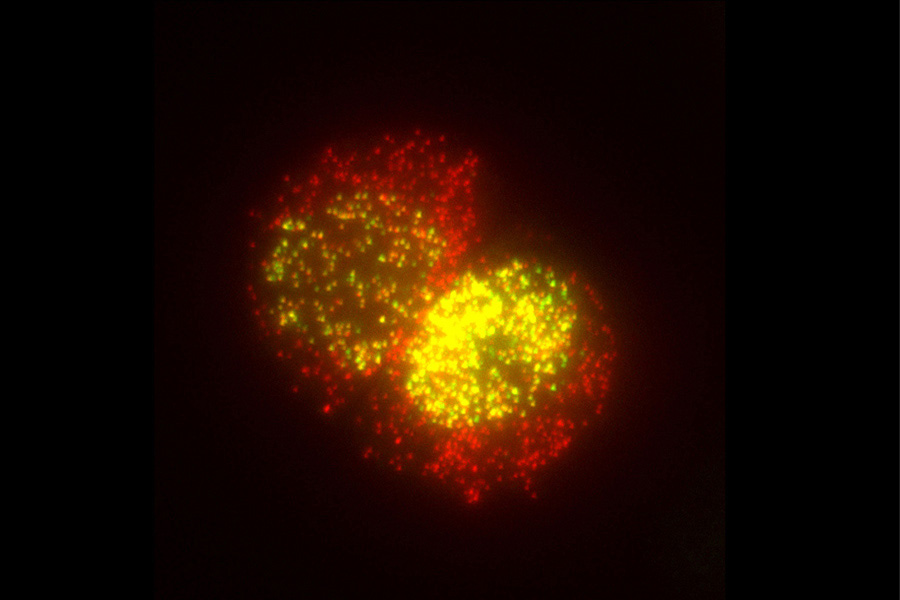
Stochastic mRNA Synthesis. With our ability to detect single mRNA molecules, we counted all mRNA molecules produced in individual cells by a gene. We found that cells of higher eukaryotes display massive cell-to-cell variations in gene expression. The variations occur because the mRNAs are produced in randomly initiated bursts and then decay in a steady manner. The observation of these bursts in mRNA synthesis raises questions about how cells are able to maintain their relatively constant phenotypes in the face of such large-scale fluctuations.
Intracellular Venues of mRNA splicing. To image individual molecules of pre-mRNA and splicing products, two sets of probes were used at the same time, one set for an intron and the other set, labeled with a different color, for an exon. Usually splicing occurs at the gene locus, while the pre-mRNA is still being synthesized. However, with our single-molecule imaging approach we found that during alternative splicing regulated by RNA binding proteins, the normally tight coupling of transcription and splicing is broken, and the pre-mRNAs are spliced in the nucleoplasm after their release from the gene locus.
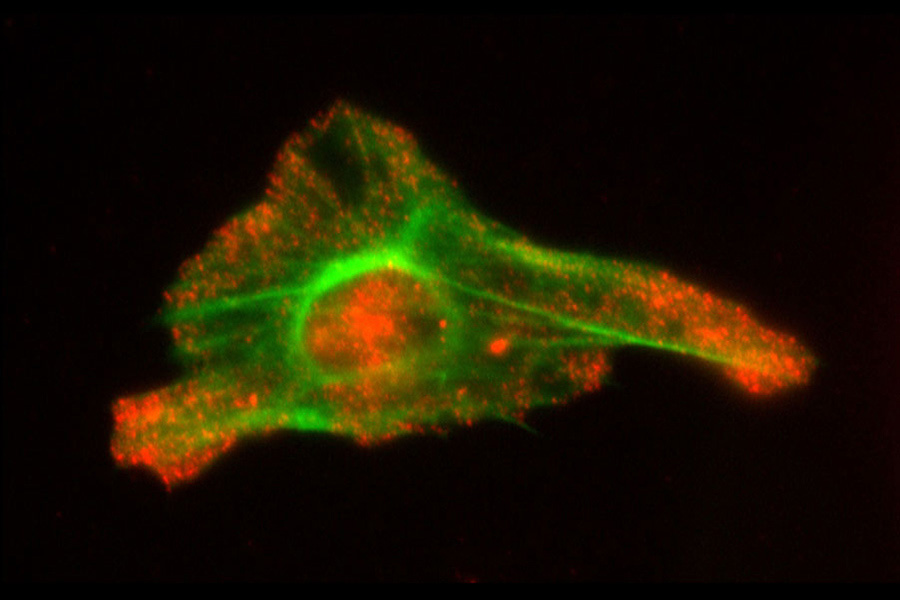
Neuronal mRNA Transport. We demonstrated that neuronal mRNAs travel into dendrites as the solitary cargo of the RNA transport granules. Our single-molecule imaging indicated that two molecules of the same or different mRNA species do not assemble in common granules. This model of mRNA transport affords a finer control of mRNA content within a synapse for synaptic plasticity then previous models did.
The current work is focused on understanding the mechanisms of mRNA transport, transcriptional bursts and on imaging other mRNA processing events like non-sense mediated decay.
Jaijyan DK, Yang S, Ramasamy S, Gu A, Zeng M, Subbian S, Tyagi S, Zhu H (2024) Imaging and quantification of human and viral circular RNAs. Nucleic Acids Res. PMI: 39051561
Kumar R, Kolloli A, Subbian S, Kaushal D, Shi L, Tyagi S (2024) Imaging the Architecture of Granulomas Induced by Mycobacterium tuberculosis Infection with Single-molecule Fluorescence In Situ Hybridization. J Immunol. PMI: 38912840
Ebraham L, Xu C, Wang A, Hernandez C, Siclari N, Rajah D, Walter L, Marras SAE, Tyagi S, Fine DH, Daep CA, Chang TL (2023) Oral epithelial cells expressing low or undetectable levels of human angiotensin-converting enzyme 2 are susceptible to SARS-CoV-2 virus infection in vitro. Pathogens 12: 843.
Jalloh S, Olejnik J, Berrigan J, Nisa A, Suder EL, Akiyama H, Lei M, Ramaswamy S, Tyagi S, Bushkin Y, Muhlberger E, Gummuluru S (2022) CD169-mediated restrictive SARS-CoV-2 infection of macrophages induces pro-inflammatory responses. PLoS Pathog 18: e1010479. PMI: 36279285
Jiang Q, Qiu Y, Kurland IJ, Drlica K, Subbian S, Tyagi S, Shi L (2022) Glutamine Is Required for M1-like Polarization of Macrophages in Response to Mycobacterium tuberculosis Infection. mBio 13: e0127422. PMI: 35762591
Bruiners N, Guerrini V, Ukey R, Dikdan RJ, Yang JH, Mishra PK, Onyuka A, Handler D, Vieth J, Carayannopoulos M, Guo S, Pollen M, Pinter A, Tyagi S, Feingold D, Philipp C, Libutti SK, Gennaro ML (2022) Longitudinal Analysis of Biologic Correlates of COVID-19 Resolution: Case Report. Front Med (Lausanne) 9: 915367. PMI: 35783607
Dikdan RJ, Marras SAE, Field AP, Brownlee A, Cironi A, Hill DA, Tyagi S (2022) Multiplex PCR Assays for Identifying all Major Severe Acute Respiratory Syndrome Coronavirus 2 Variants. Journal of Molecular Diagnostics 24: 309-319. PMI: 35121139
Yang S, Zhou H, Liu M, Jaijyan D, Cruz-Cosme R, Ramasamy S, Subbian S, Liu D, Xu J, Niu X, Li Y, Xiao L, Tyagi S, Wang Q, Zhu H, Tang Q (2022) SARS-CoV-2, SARS-CoV, and MERS-CoV encode circular RNAs of spliceosome-independent origin. J Med Virol 94: 3203-3222. PMI: 35318674
Yang S, Liu X, Wang M, Cao D, Jaijyan DK, Enescu N, Liu J, Wu S, Wang S, Sun W, Xiao L, Gu A, Li Y, Zhou H, Tyagi S, Wu J, Tang Q, Zhu H (2022) Circular RNAs Represent a Novel Class of Human Cytomegalovirus Transcripts. Microbiol Spectr: e0110622. PMI: 35604147
Vargas DY, Tyagi S, Marras SAE, Moerzinger P, Abin-Carriquiry JA, Cuello M, Rodriguez C, Martinez A, Makhnin A, Farina A, Patel C, Chuang TL, Li BT, Kramer FR (2022) Multiplex SuperSelective PCR Assays for the Detection and Quantitation of Rare Somatic Mutations in Liquid Biopsies. Journal of Molecular Diagnostics 24: 189-204. PMI: 34954118
Fraser LCR, Dikdan RJ, Dey S, Singh A, Tyagi S (2021) Reduction in gene expression noise by targeted increase in accessibility at gene loci. Proc Natl Acad Sci U S A 118. PMI: 34625470
Oliveira GP, Jr., Zigon E, Rogers G, Davodian D, Lu S, Jovanovic-Talisman T, Jones J, Tigges J, Tyagi S, Ghiran IC (2020) Detection of Extracellular Vesicle RNA Using Molecular Beacons. iScience 23: 100782. PMI: 31958756
Kumar R, Singh P, Kolloli A, Shi L, Bushkin Y, Tyagi S, Subbian S (2019) Immunometabolism of Phagocytes During Mycobacterium tuberculosis Infection. Front Mol Biosci 6: 105. PMI: 31681793
Batish M, Tyagi S (2019) Fluorescence In Situ Imaging of Dendritic RNAs at Single-Molecule Resolution. Curr Protoc Neurosci 89: e79. PMI: 31532916
Marras SAE, Bushkin Y, Tyagi S (2019) High-fidelity amplified FISH for the detection and allelic discrimination of single mRNA molecules. Proceedings of the National Academy of Sciences USA 128: 13921–13926. PMI: 31221755 https://doi.org/10.1073/pnas.1814463116
Shi L, Jiang Q, Bushkin Y, Subbian S, Tyagi S (2019) Biphasic Dynamics of Macrophage Immunometabolism during Mycobacterium tuberculosis Infection. MBio 10. PMI: 30914513
Marras SAE, Tyagi S, Antson D, Kramer FR (2019) Color-coded molecular beacons for multiplex PCR screening assays. PLoS ONE 14: e0213906
Vargas DY, Marras SAE, Tyagi S, Kramer FR (2018) Suppression of Wild-Type Amplification by Selectivity Enhancing Agents in PCR Assays that Utilize SuperSelective Primers for the Detection of Rare Somatic Mutations. J Mol Diagn 20: 415-427. PMI: 29698835
Arrigucci R, Bushkin Y, Radford F, Lakehal K, Vir P, Pine R, Martin D, Sugarman J, Zhao Y, Yap GS, Lardizabal AA, Tyagi S, Gennaro ML (2017) FISH-Flow, a protocol for the concurrent detection of mRNA and protein in single cells using fluorescence in situ hybridization and flow cytometry. Nat Protoc 12: 1245-1260. PMI: 28518171
Vargas DY, Kramer FR, Tyagi S, Marras SAE (2016) Multiplex real-time PCR assays that measure the abundance of extremely rare mutations associated with cancer. PLoS One 11: e0156546. PMI: 27244445
Radford F, Tyagi S, Gennaro ML, Pine R, Bushkin Y (2016) Flow cytometric characterization of antigen-specific T cells based on RNA and its advantages in detecting infections and immunological disorders. Critical Reviews in Immunology 5: 359-378. PMI:
Vir P, Arrigucci R, Lakehal K, Davidow AL, Pine R, Tyagi S, Bushkin Y, Lardizabal A, Gennaro ML (2015) Single-Cell Cytokine Gene Expression in Peripheral Blood Cells Correlates with Latent Tuberculosis Status. PLoS One 10: e0144904. PMI: 26658491
Tyagi S (2015) Tuning noise in gene expression. Mol Syst Biol 11: 805. PMI:
Patil S, Fribourg M, Ge Y, Batish M, Tyagi S, Hayot F, Sealfon SC (2015) Single-cell analysis shows that paracrine signaling by first responder cells shapes the interferon-beta response to viral infection. Sci Signal 8: ra16. PMI: 25670204
Bushkin Y, Radford F, Pine R, Lardizabal A, Mangura BT, Gennaro ML, Tyagi S (2015) Profiling T cell activation using single-molecule fluorescence in situ hybridization and flow cytometry. J Immunol 194: 836-841. PMI: 25505292
Bijlard M, Klunder B, de Jonge JC, Nomden A, Tyagi S, de Vries H, Hoekstra D, Baron W (2015) Transcriptional expression of myelin basic protein in oligodendrocytes depends on functional syntaxin 4: a potential correlation with autocrine signaling. Mol Cell Biol 35: 675-687. PMI: 25512606
Xu S, Tyagi S, Schedl P (2014) Spermatid cyst polarization in Drosophila depends upon apkc and the CPEB family translational regulator orb2. PLoS Genet 10: e1004380. PMI: 24830287
Markey FB, Ruezinsky W, Tyagi S, Batish M (2014) Fusion FISH imaging: single-molecule detection of gene fusion transcripts in situ. PLoS One 9: e93488. PMI: 24675777
Shah K, Tyagi S (2013) Barriers to transmission of transcriptional noise in a c-fos c-jun pathway. Mol Syst Biol 9: 687. PMI: 24022005
Tyagi S, Kramer FR (2012) Molecular beacons in diagnostics. Faculty of 1000 Medicine Reports 4: 4-10. PMI:
Batish M, van den Bogaard P, Kramer FR, Tyagi S (2012) Neuronal mRNAs travel singly into dendrites. Proc Natl Acad Sci USA 109: 4645-4650. PMI: 22392993
Vargas DY, Shah K, Batish M, Levandoski M, Sinha S, Marras SA, Schedl P, Tyagi S (2011) Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell 147: 1054-1065. PMI: 22118462
Tyagi S (2010) E. coli, what a noisy bug. Science 329: 518-519. PMI: 20671174
Tyagi S (2009) Imaging intracellular RNA distribution and dynamics in living cells. Nat Methods 6: 331-338. PMI: 19404252
Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S (2008) Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5: 877-879. PMI: 18806792
Tyagi S (2007) RT-PCR enters the realm of stochastic gene expression. Genetic Engineering News: March 2007.: PMI:
Tyagi S (2007) Splitting or stacking fluorescent proteins to visualize mRNA in living cells. Nat Methods 4: 391-392. PMI: 17464293
Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S (2006) Stochastic mRNA synthesis in mammalian cells. PLoS Biol 4: e309. PMI: 17048983
Vargas DY, Raj A, Marras SAE, Kramer FR, Tyagi S (2005) Mechanism of mRNA transport in the nucleus. Proc Natl Acad Sci USA 102: 17008-17013. PMI: 16284251
Mhlanga MM, Vargas DY, Fung CW, Kramer FR, Tyagi S (2005) tRNA-linked molecular beacons for imaging mRNAs in the cytoplasm of living cells. Nucleic Acids Res 33: 1902-1912. PMI: 15809226
Tyagi S, Alsmadi O (2004) Imaging native beta-actin mRNA in motile fibroblasts. Biophys J 87: 4153-4162. PMI: 15377515
Bratu DP, Cha BJ, Mhlanga MM, Kramer FR, Tyagi S (2003) Visualizing the distribution and transport of mRNAs in living cells. Proc Natl Acad Sci USA 100: 13308-13313. PMI: 14583593
Marras SAE, Kramer FR, Tyagi S (2002) Efficiencies of fluorescence resonance energy transfer and contact-mediated quenching in oligonucleotide probes. Nucleic Acids Research 30: e122. PMI: 12409481
Tyagi S, Marras SAE, Kramer FR (2000) Wavelength-shifting molecular beacons. Nature Biotechnology 18: 1191-1196.: PMI: 11062440
Marras SAE, Kramer FR, Tyagi S (1999) Multiplex detection of single-nucleotide variations using molecular beacons. Genetic Analysis: Biomolecular Engineering 14: 151-156.: PMI: 10084107
Bonnet G, Tyagi S, Libchaber A, Kramer FR (1999) Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc Natl Acad Sci USA 96: 6171-6176. PMI: 10339560
Tyagi S, Bratu DP, Kramer FR (1998) Multicolor molecular beacons for allele discrimination. Nat Biotechnol 16: 49-53. PMI: 9447593
Piatek AS, Tyagi S, Pol AC, Telenti A, Miller LP, Kramer FR, Alland D (1998) Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat Biotechnol 16: 359-363. PMI: 9555727
Kostrikis LG, Tyagi S, Mhlanga MM, Ho DD, Kramer FR (1998) Spectral genotyping of human alleles. Science 279: 1228-1229. PMI: 9508692
Tyagi S, Landegren U, Tazi M, Lizardi PM, Kramer FR (1996) Extremely sensitive, background-free gene detection using binary probes and beta replicase. Proc Natl Acad Sci USA 93: 5395-5400. PMI: 8643586
Tyagi S, Kramer FR (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol 14: 303-308. PMI: 9630890
Tyagi S, Ponnamperuma C (1990) Nonrandomness in prebiotic peptide synthesis. J Mol Evol 30: 391-399. PMI: 2111852
Lomeli H, Tyagi S, Pritchard CG, Lizardi PM, Kramer FR (1989) Quantitative assays based on the use of replicatable hybridization probes. Clin Chem 35: 1826-1831. PMI: 2673578
Tyagi S (1981) Origins of translation: the hypothesis of permanently attached adaptors. Orig Life 11: 343-351. PMI: 6799890