Salvatore A.E. Marras, Ph.D.
Assistant Professor of Microbiology, Biochemistry & Molecular Genetics
marrassa@njms.rutgers.edu | Laboratory Website
+1-973-854-3373
room W350-A
NEWS
Our latest publications:
Ma MT, Jiang Q, Chen CH, Badeti S, Wang X, Zeng C, Evans D, Bodnar B, Marras SAE, Tyagi S, Bharaj P, Yehia G, Romanienko P, Hu W, Liu SL, Shi L, Liu D (2024) S309-CAR-NK cells bind the Omicron variants in vitro and reduce SARS-CoV-2 viral loads in humanized ACE2-NSG mice. Journal of Virology: e0003824. PMI: 38767356. To view and download the article, visit www.journals.asm.org
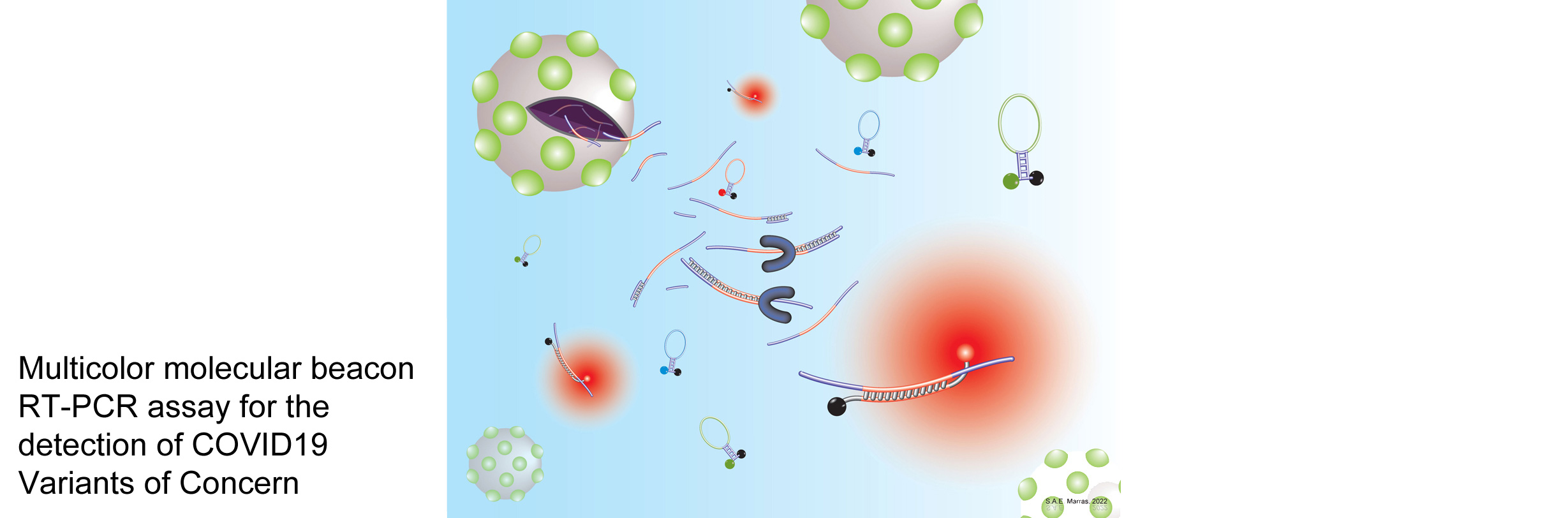
Banada PP, Green R, Streck D, Kurvathi R, Reiss R, Banik S, Daivaa N, Montalvan I, Jones R, Marras SAE, Chakravorty S, Alland D (2023) An expanded RT-PCR melting temperature coding assay to rapidly identify all known SARS-CoV-2 variants and sub-variants of concern. Scientific Reports 13: 21927. PMI: 38081834. To view and download the article, visit www.nature.com
Ebraham L, Xu C, Wang A, Hernandez C, Siclari N, Rajah D, Walter L, Marras SAE, Tyagi S, Fine DH, Daep CA, Chang TL (2023) Oral epithelial cells expressing low or undetectable levels of human angiotensin-converting enzyme 2 are susceptible to SARS-CoV-2 virus infection in vitro. Pathogens 12: 843. To view and download the article, visit www.mdpi.com
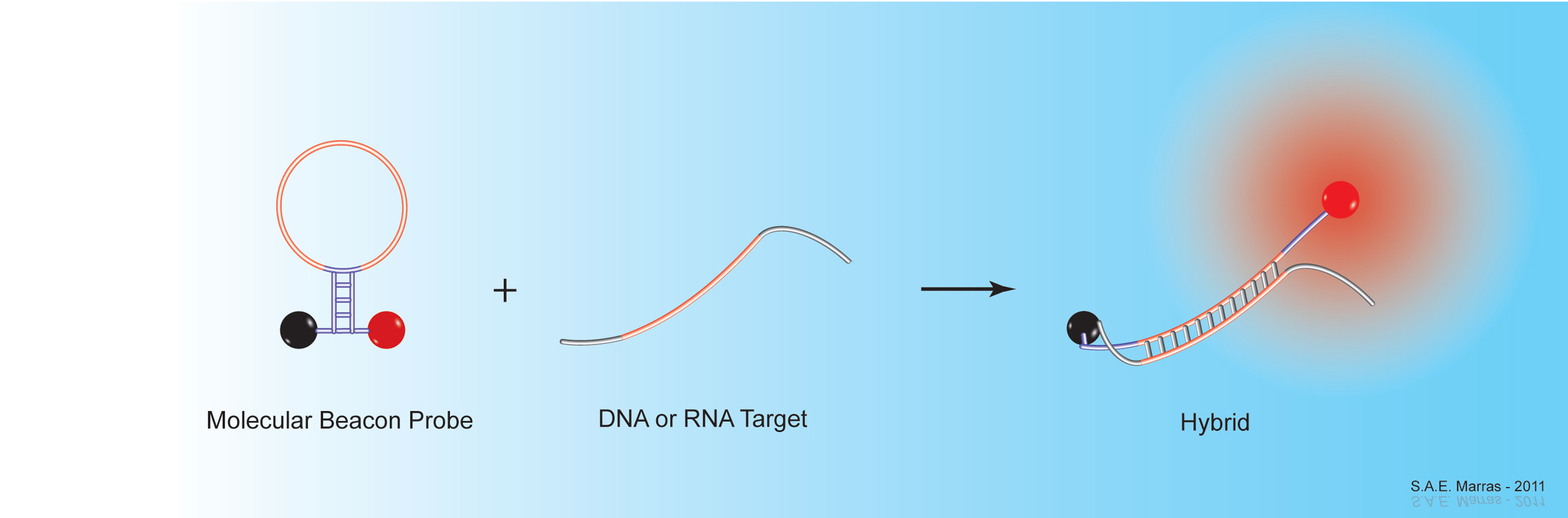
The introduction of self-quenching fluorescent nucleic acid hybridization probes has markedly improved the ability to detect RNA and DNA sequences with high sensitivity and specificity. Self-quenching probes are useful in situations where it is not possible or desirable to separate probes that are not hybridized to target sequences from probes that are hybridized to target sequences. These probes are used for the real-time monitoring of nucleic acid amplification assays, such as polymerase chain reactions (PCR) and nucleic acid sequence-based amplification (NASBA) assays. Real-time amplification enables the detection and quantitative measurement of rare DNA and RNA targets in clinical samples. Furthermore, these assays can be carried out in sealed reaction tubes, thereby preventing the contamination of untested samples. Since fluorescent nucleic acid hybridization probes remain dark when not hybridized to a target sequence, they also enable the detection of DNA and RNA targets in living cells.
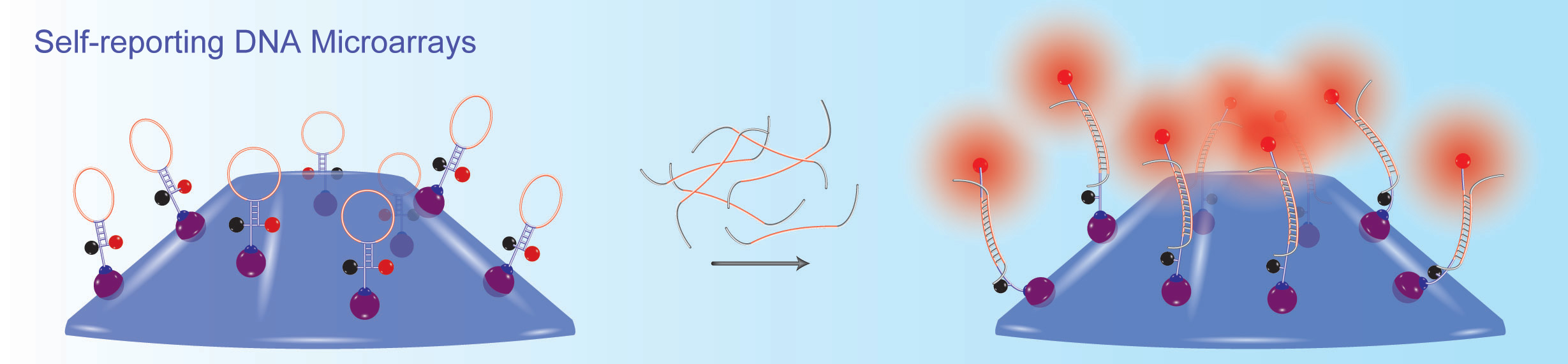
 Together with his colleagues, Sanjay Tyagi and Fred Russell Kramer, Salvatore Marras developed molecular beacons, one of the first fluorescent nucleic acid hybridization probe technologies. His research focuses on the different properties of fluorescent hybridization probes, such as design parameters that affect their specificity, and on the effects of interactions between different fluorophore & quencher pairs. His group also develops novel nucleic acid detection methods, including: self-reporting DNA microarray platforms; highly multiplexed, real-time nucleic acid amplification assays for the detection of infectious agents in clinical samples; and extremely sensitive in situ and in vivo hybridization methods, utilizing organic-based fluorescent reporters and metal-based luminescent compounds.
Together with his colleagues, Sanjay Tyagi and Fred Russell Kramer, Salvatore Marras developed molecular beacons, one of the first fluorescent nucleic acid hybridization probe technologies. His research focuses on the different properties of fluorescent hybridization probes, such as design parameters that affect their specificity, and on the effects of interactions between different fluorophore & quencher pairs. His group also develops novel nucleic acid detection methods, including: self-reporting DNA microarray platforms; highly multiplexed, real-time nucleic acid amplification assays for the detection of infectious agents in clinical samples; and extremely sensitive in situ and in vivo hybridization methods, utilizing organic-based fluorescent reporters and metal-based luminescent compounds.
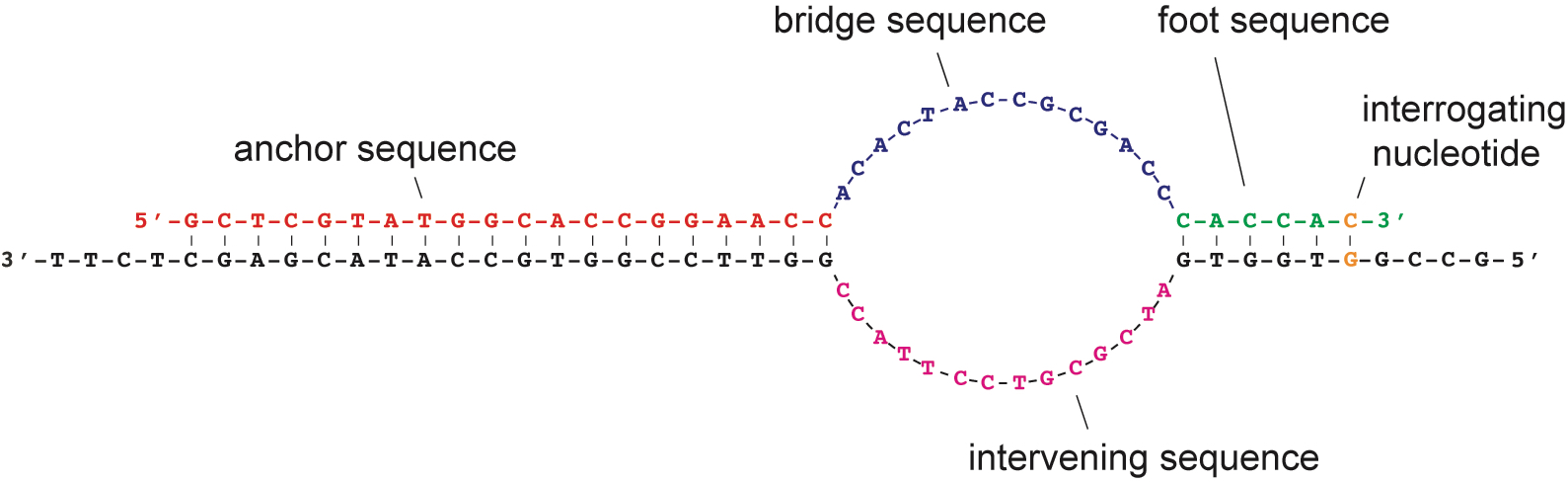
More recently, Dr. Marras co-developed the SuperSelective primer technology, which enables the detection and quantitation of somatic mutations whose presence relates to cancer diagnosis, prognosis, and therapy, in real-time multiplex PCR assays that can potentially analyze rare DNA fragments present in blood samples (liquid biopsies). However, SuperSelective primer applications are not limited to cancer diagnosis as they can be used to identify and quantify any rare mutant DNA fragment in a large background of wild-type DNA fragments, such as the detection and identification of heteroresistant bacteria and fungi species.
See a complete bibliography on PubMed
Banada PP, Green R, Streck D, Kurvathi R, Reiss R, Banik S, Daivaa N, Montalvan I, Jones R, Marras SAE, Chakravorty S, Alland D (2023) An expanded RT-PCR melting temperature coding assay to rapidly identify all known SARS-CoV-2 variants and sub-variants of concern. Scientific Reports 13: 21927. PMI: 38081834
Ebraham L, Xu C, Wang A, Hernandez C, Siclari N, Rajah D, Walter L, Marras SAE, Tyagi S, Fine DH, Daep CA, Chang TL (2023) Oral epithelial cells expressing low or undetectable levels of human angiotensin-converting enzyme 2 are susceptible to SARS-CoV-2 virus infection in vitro. Pathogens 12: 843.
Narang A, Marras SAE, Kurepina N, Chauhan V, Shashkina E, Kreiswirth B, Varma-Basil M, Vinnard C, Subbian S (2022) Ultrasensitive detection of multidrug-resistant Mycobacterium tuberculosis using SuperSelective primer-based real-time PCR assays. International Journal of Molecular Sciences 23(24): 1575223. PMI: 36555395
Badeti S, Jiang Q, Naghizadeh A, Tseng HC, Bushkin Y, Marras SAE, Nisa A, Tyagi S, Chen F, Romanienko P, Yehia G, Evans D, Lopez-Gonzalez M, Alland D, Russo R, Gause W, Shi L, Liu D (2022) Development of a novel human CD147 knock-in NSG mouse model to test SARS-CoV-2 viral infection. Cell & Bioscience 12: 88. PMI: 35690792
Dikdan RJ, Marras SAE, Field AP, Brownlee A, Cironi A, Hill DA, Tyagi S (2022) A multiplex PCR assay for identifying all major SARS-CoV-2 variants. Journal of Molecular Diagnostics 24: 309-319. PMI: 35121139
Vargas DY, Tyagi S, Marras SAE, Moerzinger P, Abin-Carriquiry JA, Cuello M, Rodriguez C, Martinez A, Makhnin A, Farina A, Patel C, Chuang TL, Li BT, Kramer FR (2022) Multiplex SuperSelective PCR assays for the detection and quantitation of rare somatic mutations in liquid biopsies. Journal of Molecular Diagnostics 24: 189-204. PMI: 34954118
Marras SAE, Chen L, Shashkina E, Davidson RM, Strong M, Daley CL, Kreiswirth BN (2021) A molecular-beacon-based multiplex real-time PCR assay to distinguish Mycobacterium abscessus subspecies and determine macrolide susceptibility. Journal of Clinical Microbiology 59: e0045521. PMI: 33980653
Whitfield MG, Marras SAE, Warren RM, Van Rie A, Rice J, Wangh LJ, Kreiswirth BN (2020) Rapid Pyrazinamide drug susceptibility testing using a closed-tube PCR assay of the entire pncA gene. Scientific Reports 10: 4234. PMI: 32144379
Marras SAE, Bushkin Y, Tyagi S (2019) High-fidelity amplified FISH for the detection and allelic discrimination of single mRNA molecules. Proceedings of the National Academy of Sciences USA 128: 13921–13926. PMI: 31221755
Marras SAE, Tyagi S, Antson DO, Kramer FR (2019) Color-coded molecular beacons for multiplex PCR screening assays. PLoS One 14: e0213906. PMI: 30883590
Vargas DY, Marras SAE, Tyagi S, Kramer FR (2018) Suppression of wild-type amplification by selectivity enhancing agents in PCR assays that utilize SuperSelective primers for the detection of rare somatic mutations. Journal of Molecular Diagnostics 20: 415-427. PMI: 29698835
Vargas DY, Kramer FR, Tyagi S, Marras SAE (2016) Multiplex real-time PCR assays that measure the abundance of extremely rare mutations associated with cancer. PLoS One 11: e0156546. PMI: 27244445
Wirpsza L, Pillai S, Batish M, Marras SAE, Krasnoperov L, Mustaev A (2012) Highly bright avidin-based affinity probes carrying multiple lanthanide chelates. Journal of Photochemistry and Photobiology B: Biology 116: 22-29. PMI: 23018156
Catrina IE, Marras SAE, Bratu DP (2012) Tiny molecular beacons: LNA/2′-O-methyl RNA chimeric probes for imaging dynamic mRNA processes in living cells. ACS Chemical Biology 7: 1586-1595. PMI: 22738327
Dai A, Yang W, Firlar E, Marras SAE, Libera M (2012) Surface-patterned microgel-tethered molecular beacons. Soft Matter 8: 3067-3076. PMI: 0
Vargas DY, Shah K, Batish M, Levandoski M, Sinha S, Marras SAE, Schedl P, Tyagi S (2011) Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell 147: 1054-1065. PMI: 22118462
Chakravorty S, Aladegbami B, Burday M, Levi M, Marras SAE, Shah D, El-Hajj HH, Kramer FR, Alland D (2010) Rapid universal diagnosis of bacterial pathogens from clinical cultures using a novel sloppy molecular beacon melting temperature signature technique. Journal of Clinical Microbiology 48: 258-267. PMI: 19923485
El-Hajj HH, Marras SAE, Tyagi S, Shashkina E, Kamboj M, Kiehn TE, Glickman MS, Kramer FR, Alland D (2009) Use of sloppy molecular beacon probes for identification of mycobacterial species. Journal of Clinical Microbiology 47: 1190-1198. PMI: 19171684
Marras SAE (2008) Interactive fluorophore and quencher pairs for labeling fluorescent nucleic acid hybridization probes. Molecular Biotechnology 38: 247-255. PMI: 17985254
Marras SAE, Tyagi S, Kramer FR (2006) Real-time assays with molecular beacons and other fluorescent nucleic acid hybridization probes. Clinica Chimica Acta 363: 48-60. PMI: 16111667
Vargas DY, Raj A, Marras SAE, Kramer FR, Tyagi S (2005) Mechanism of mRNA transport in the nucleus. Proceedings of the National Academy of Sciences USA 102: 17008-17013. PMI: 16284251
Marras SAE, Kramer FR, Tyagi S (2002) Efficiencies of fluorescence resonance energy transfer and contact-mediated quenching in oligonucleotide probes. Nucleic Acids Research 30: e122. PMI: 12409481
El-Hajj HH, Marras SAE, Tyagi S, Kramer FR, Alland D (2001) Detection of rifampin resistance in Mycobacterium tuberculosis in a single tube with molecular beacons. Journal of Clinical Microbiology 39: 4131-4137.: PMI: 11682541
Tyagi S, Marras SAE, Kramer FR (2000) Wavelength-shifting molecular beacons. Nature Biotechnology 18: 1191-1196.: PMI: 11062440
Vet JAM, Majithia AR, Marras SAE, Tyagi S, Dube S, Poiesz BJ, Kramer FR (1999) Multiplex detection of four pathogenic retroviruses using molecular beacons. Proceedings of the National Academy of Sciences USA 96: 6394-6399.: PMI: 10339598
Marras SAE, Kramer FR, Tyagi S (1999) Multiplex detection of single-nucleotide variations using molecular beacons. Genetic Analysis: Biomolecular Engineering 14: 151-156.: PMI: 10084107